Application & Tech Notes

TITLE
Single Cell Sequencing Tumor Infiltrating Lymphocytes Lung Cancer
INTRODUCTION
In the realm of cancer immunotherapy, understanding the complex interactions between tumors and the immune system is crucial for devising effective treatment strategies. Tumor-infiltrating lymphocytes (TILs) play a pivotal role in the anti-tumor immune response and can serve as valuable indicators of patient prognosis and therapeutic response. The advent of advanced single-cell sequencing technologies has revolutionized the field by enabling comprehensive profiling of TIL populations. Here, we demonstrate the use of the SingulatorTM Platform to generate high quality single-cell suspensions from normal and tumor human lung tissue with up to 94% viability. These suspensions were used as input to the 10x Single Cell 3’ v3.1 Gene Expression Assay and to purify TILs from the tumor samples using immunogenic bead separation, demonstrating the epitopes remain intact on the surface of the TILs through the SingulatorTM process. Finally, we characterized the gene expression differences between matched normal lung tissues, tumor samples and TILs from 3 patients and identified a gene expression profile that is specific to the TILs purified from the tumor samples.

TITLE
Dissociating Tumor Tissue into Cells with S2 Genomics’ Pan Tumor Reagent and the Singulator Platform
INTRODUCTION
Cancer is a complex disease that arises from alterations in the genetic material of cells, leading to uncontrolled growth and proliferation. Tumor sequencing has emerged as a powerful tool for understanding the genetic changes that occur in cancer cells and identifying potential therapeutic targets. However, tumors are heterogeneous, consisting of different types of cells with varying genetic profiles. Therefore, it is essential to dissociate tumors into the constituent cells and perform sequencing at the single cell level to obtain accurate information about the genetic changes driving the cancer. The conventional techniques for dissociating solid tumors are often intricate, require considerable time and expertise, and yield variable results. In this study, we showcase the efficacy of the Singulator™ platform as a means of isolating viable lung tumor cells in high yield and viability from patients diagnosed with adenocarcinoma and squamous cell carcinoma. Our approach incorporates the utilization of S2 Genomics’ Pan Tumor Reagent, which enables robust and reliable tumor cell isolation.

TITLE
Isolation of Viable Cells from Mouse Heart Tissue with the Singulator™ 100
INTRODUCTION
Research into cardiovascular diseases can benefit greatly from single-cell approaches to genomics and cell biology analyses. However, traditional methods for viable cell isolations from heart tissue are complex, time-consuming, and technically challenging, especially so for cardiomyocytes due to their fragility. Single-cell sequencing libraries from heart cells are also difficult to generate on droplet-based microfluidics platforms due to cell size restrictions. Here, we demonstrate the use of the Singulator™ 100 for the isolation of viable mouse heart cells with high yields, using S2 Genomics’ Heart Cell Reagent and Large Cell Isolation Cartridge. In addition, we have connected the workflow to Parse Biosciences’ Evercode WT v2 kit for generation of scRNA-Seq data, showing that the Parse technology can be used to generate sequencing libraries with cardiomyocyte cell-type representation.

TITLE
Isolation of Mouse Skin Cells with the Singulator™ Platform
INTRODUCTION
The skin is the outermost protective barrier organism and comprises of two main layers, the epidermis and the dermis. To understand skin homeostasis in healthy and diseased states, a thorough molecular characterization of every type of skin cell type is required. This in turn requires isolation of viable cells from skin for downstream genomics and cell biology analyses. However, traditional methods for isolating cells from skin tissue, are time-consuming, complex, and technically challenging. Here, we demonstrate the use of the Singulator™ 100 for isolation of viable mouse skin cells with high yield with S2 Genomics’ Skin Cell Reagent.

TITLE
Sequencing Skin and Kidney Cells: Leveraging the Singulator™ 100 and Parse Biosciences’ Evercode™ Whole Transcriptome v2 Platform
INTRODUCTION
Advancements in high-throughput sequencing technologies have revolutionized our understanding of cellular heterogeneity and gene expression patterns. To comprehensively investigate the transcriptional landscapes of specific cell types within complex tissues, researchers have turned to single-cell RNA sequencing (scRNA-seq) techniques. The availability of multiple platforms for single-cell sequencing is important to allow researchers to select the appropriate technology to address a specific scientific question. In this technical note, we demonstrate the compatibility of Singulator-produced skin and kidney cells with the Parse Biosciences single-cell whole transcriptome technology. The ability of the Evercode technology to both fix cells and process many samples in parallel facilitates a wider range of sequencing applications when combined with the ease of use and reproducibility of the Singulator system.

TITLE
Sequencing Lung and Kidney Nuclei: Leveraging the Singulator™ 100 and Parse Biosciences Evercode™ Whole Transcriptome v2 Platform
INTRODUCTION
Advancements in high-throughput sequencing technologies have revolutionized our understanding of cellular heterogeneity and gene expression patterns. To comprehensively investigate the transcriptional landscapes of specific cell types within complex tissues, researchers have turned to single nucleus RNA sequencing (snRNA-seq) techniques. In this technical note, we demonstrate the compatibility of Singulator produced kidney and lung nuclei with the application of the Parse Biosciences single-cell whole transcriptome technology. The utilization of multiple platforms for single nuclei sequencing is crucial as it allows researchers to overcome the limitations and biases associated with individual technologies. The ability to both fix and run multiple samples simultaneously simplifies the single nuclei sequencing workflow, and combined with the ease of the Singulator system facilitates a wider range of sequencing applications.

TITLE
Use of the S2 Genomics Nuclei Debris Removal Stock Reagent
INTRODUCTION
Single-cell and single-nucleus sequencing techniques have transformed our ability to study the genomics of individual cells, providing insights into the diversity and heterogeneity of biological systems. However, the quality of the cells or nuclei in suspension used to generate a sequencing library can significantly affect the results of these experiments, and in particular, the presence of intracellular and extracellular debris in the sample can have significant adverse effects. The debris can negatively impact the degree of mitochondrial contamination and overall sequencing library quality (genes detected, unique UMIs, percentage of reads mapped to cells, etc). Debris can also lead to clogging of microfluidic channels, high ambient RNA, and background noise leading to diminished data quality. Brain cell or nuclei preparations, for example, can contain high amounts of myelin debris that must be removed, and liver samples may contain high amounts of intracellular debris from hepatocytes containing mitochondria and cellular remnants. To address this issue, we have developed a Nuclei Debris Removal Stock Reagent (100-253-628) that improves the quality of nuclei suspensions prior to sequencing library generation. In this technical note, we describe use of the reagent for cleaning intracellular and extracellular debris from mouse brain and liver nuclei preparations.

TITLE
Use of NIC+ with the Singulator™ 100 For Sub-20 mg Samples
ABSTRACT
The Singulator™ 100 can be used to quickly and efficiently isolate nuclei from a wide variety of tissue types, including samples that are fresh, frozen, or OCT preserved. It is ideal for genomics, cell biology and other ‘omics applications, including scRNA-Seq, snRNA-Seq, ATAC-Seq, CITE-Seq, FACS, and immuno-oncology. S2 Genomics provides a selection of customizable pre-set protocols and pre-formulated reagents for nuclei isolations from an expanding set of mouse, rat, and human tissues, including tumors. The new NIC+™ sample cartridge for nuclei isolation extends the range of sample sizes compatible with dissociation on the Singulator down to 1 mg. Here we describe the performance of the NIC+ cartridges and the Singulator 100 for nuclei isolations from samples below 20 mg in size.

TITLE
Use of RNase Inhibitor with the Singulator™ 100
ABSTRACT
The Singulator™ 100 can be used to quickly and efficiently isolate nuclei from a wide variety of tissue types, and from samples that are fresh, frozen, or OCT preserved. If the nuclei are intended for downstream RNA analysis, it is critical that care be taken to prevent degradation of the RNA in the nuclei by the activity of endogenous RNase enzymes, most importantly RNase A. To help preserve RNA integrity, RNAse inhibitor is commonly added to samples during the isolation and purification of nuclei. Here we describe the use of RNase inhibitors with the Singulator for nuclei isolations.
TITLE
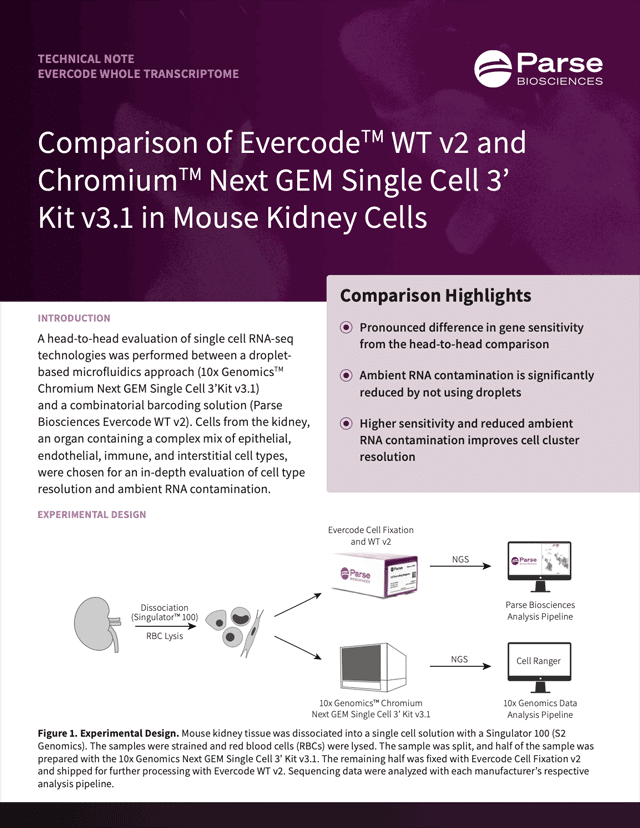
Parse Biosciences | Comparison of Evercode™ WT v2 and Chromium™ Next GEM Single Cell 3’ Kit v3.1 in Mouse Kidney Cells
ABSTRACT
A head-to-head evaluation of single cell RNA-seq technologies was performed between a droplet- based microfluidics approach (10x Genomics™ Chromium Next GEM Single Cell 3’Kit v3.1) and a combinatorial barcoding solution (Parse Biosciences Evercode WT v2). Cells from the kidney, an organ containing a complex mix of epithelial, endothelial, immune, and interstitial cell types, were chosen for an in-depth evaluation of cell type resolution and ambient RNA contamination.
TITLE
Singulator 100™ and MARS®: Efficient Nuclei Isolation from a variety of tissue samples
ABSTRACT
MARS® acoustic technology utilizes active-microfluidics acoustics for the separation of single cells or nuclei without labeling, based only on the difference in their physical parameters. The cells are isolated with high recovery and ready for single-cell genomics.
The Singulator™ 100 System automates the processing of solid tissue samples into suspensions of single cells or nuclei with high yields and from small samples for a wide range of single-cell biology and genomic analyses.
