Unlocking Tumor Heterogeneity with Single-Cell Analysis
Single-cell analysis has revolutionized oncology research, providing detailed insights into tumor cell heterogeneity. Understanding the diversity of cell types within tumors and their genetic, epigenetic, and phenotypic variations is crucial for identifying therapeutic targets and developing effective treatments.
Central to this process is the isolation of high-quality single-cells or single-nuclei from tumor samples. This critical step can be challenging, as it requires preserving all relevant cell populations within the tissue. In this blog, we explore how thoughtful experimental design and reliable sample preparation techniques improve single-cell dissociation for cancer research, facilitating accurate characterization of tumor cell heterogeneity and enabling studies targeting specific cell populations.
Designing Experiments to Profile Tumor Heterogeneity
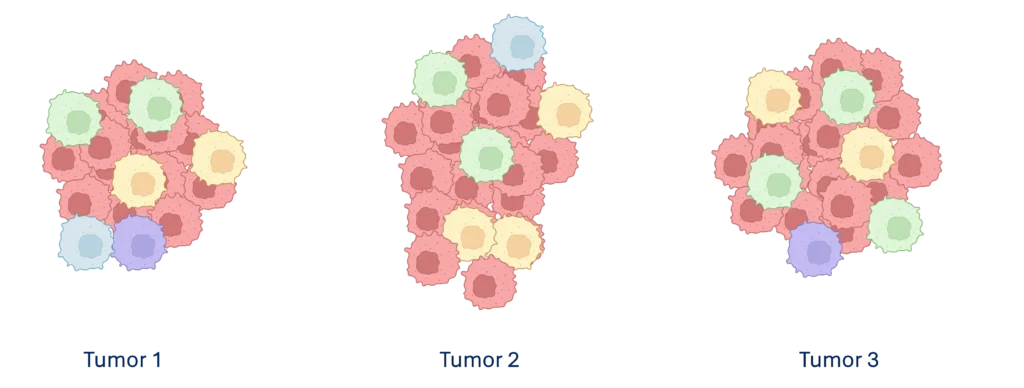
Tumor microenvironments are complex, containing diverse cell populations that vary across tissue types and even among tumors within the same patient (Figure 1). Achieving reliable research outcomes starts with thoughtful experimental design, while considering the research objectives, single-cell assay selection, and sample characteristics.
Bulk Analysis vs Single-Cell
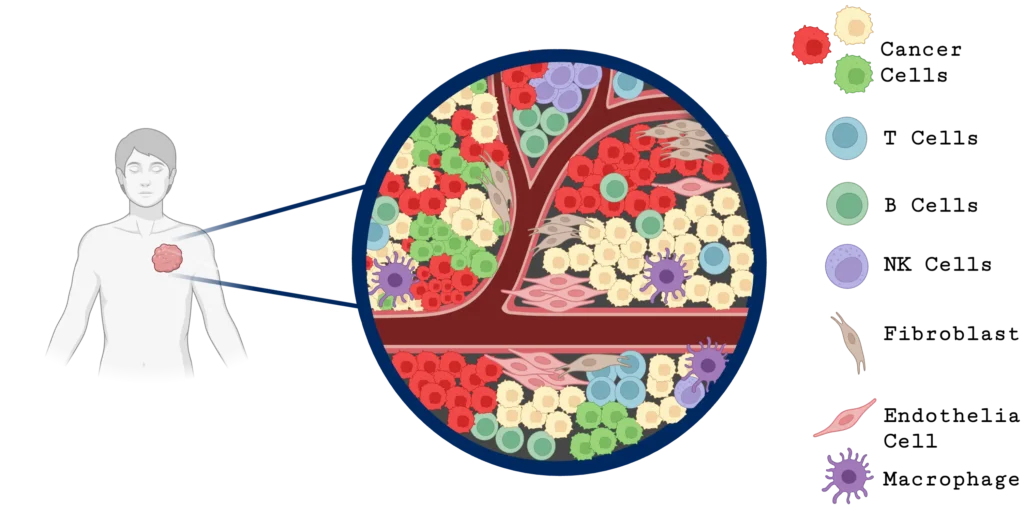
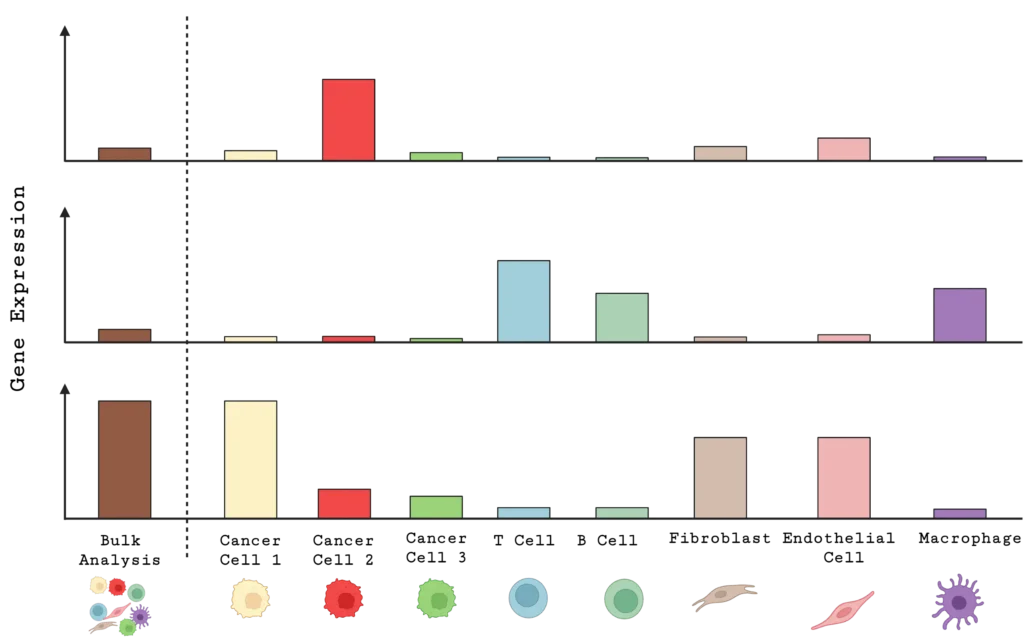
Single-cell analysis offers a significant advantage to traditional bulk gene expression analysis for studying tumor heterogeneity. While bulk analysis averages signals across all the cells in a tissue sample, potentially obscuring important cellular differences, single-cell analysis reveals changes within a chosen cell type (Figure 2). This detailed view enables researchers to identify changes in DNA sequence and structure, gene expression, and/or protein expression in specific subpopulations, such as B cells, T cells, fibroblasts, natural killer cells, endothelial cells, and macrophages, that can influence tumor behavior, therapy resistance, and immune evasion
Considerations for Sample Preparation for Single-Cell Genomics Assays
Next, let’s explore different factors that scientists need to consider when deciding between different methods for single-cell genomics experiments.
Preservation Method
The tissue preservation method can significantly impact the choice of cell isolation technique and the downstream assay. For retrospective studies, the preservation method can dictate the isolation method that scientists will need to employ. For instance, frozen and OCT-embedded tissues typically require nuclei isolation, whereas fresh tissues can be dissociated into either single cells or single nuclei. All 3 tissues are available to most single cell assays, making these preservation methods the most utilized. Recently, 10x Genomics® released the 10x Flex assay that can measure gene expression in FFPE preserved tissues. Scientist can now decide to isolate cells or nuclei from FFPE, depending on the target population. More on that later.
Tissue-Specific Challenges
Certain tissues can present unique challenges for single-cell dissociation. For example, large or fragile cell types, like hepatocytes, are better suited for nuclei isolation. In tissues with high endogenous RNase activity, like pancreas tissue,1 scientists may gain more insights from DNA sequencing or ATAC sequencing rather than mRNA profiling. Luckily, many cancer tissues don’t perform like the adjacent normal tissues and are more easily dissociated into single-nuclei, often making cancer biopsies more amenable to single-cell studies than the healthy tissue. Examples include isolating nuclei from hepatocellular carcinoma and pancreas adenocarcinoma.2
When tumors contain a high number of necrotic and apoptotic cells, those isolates may have a high percentage of dead cells and debris. Scientists should examine their single-cell suspensions under a microscope to determine if an additional clean up like FACS sorting, Levitation technology, or a negative bead selection is required. Keep in mind, these clean ups can be lossy, so it is a balance between having enough cells and a clean preparation.
Cell Type Suitability
With the proliferation of single-cell experiments and subsequent publications, a few trends have emerged in how the sample preparation method can influence the types of cells recovered, which will impact results of the experiment.
First, studies that employ fresh cell isolations typically bias their results toward hearty immune cells, such as T-cells and B-cells, while eliminating the attached cell types, like podocytes3 in kidney and hepatocytes in liver. For most tissues and cell types, isolating nuclei is more efficient at recovering attached cell types. One notable exception is tissue-resident neutrophils that are often underrepresented in both cell isolations from fresh tissues and nuclei isolations from flash frozen tissues.4
Second, in looking at FFPE tissues, we see that neutrophils can be effectively captured in either cell or nuclei preparations. Similar to the results seen from non-fixed tissues, we have seen that immune cells are overrepresented in cell isolates compared to nuclei isolates when taken from the same tumor sample. Interestingly, tissue-resident neutrophils are captured in the cell isolates, offering up a potential sample preservation method for these tough to isolate cells.
Savvy researchers who are targeting specific cell types will explore if cell isolations or nuclei isolations yield more of their cell type of interest. They may even need to explore different preservation methods, if working with exceptionally challenging tissues like pancreas or fragile cells like neutrophils.
Cell Type Selection
In addition to the preservation method and sample preparation technique having an impact on the cells types isolated, scientists can leverage enrichment methods to select for their cell types of interest, increasing the richness of their data, while reducing the cost per data point.
The first method uses cell surface markers to enrich for a population by using antibodies that specifically target proteins on the cell surface. Antibody-bound cells can then be enriched using fluorescent tags and FACS sorting or positively selected through bead-based enrichment methods. Both approaches require an antibody that is specific to the cell surface protein, and not all cell types can be targeted.
The second, more recent method is Programmable Enrichment via RNA FlowFISH by sequencing (PERFF-seq)5, a recent development from Stanford University and MSKCC. In PERFF-seq, scientists isolate nuclei from FFPE samples, which are then selected for based off their mRNA content. In this way, scientists can design probes that target any expressed gene and enrich for rare nuclei from the nuclei suspensions.
Downstream Single-Cell Assay
One final consideration is the downstream assay. In some cases, the sample isolation method is determined by the downstream assays to be performed; ATAC-Seq and multiomic studies require nuclei isolation, whereas TCR and BCR sequencing and cell surface marker detection methods, like CITE-Seq, necessitate whole cell isolation. In other cases, like FFPE preserved tissues, the preservation method will currently exclude downstream assays other than the 10x Flex kit which measures gene expression.
No One-Size-Fits-All Sample Preparation Method
As discussed, many factors can impact the preparation of cancer samples, and there is no one-size-fits-all sample preparation method for single-cell genomics assays. While we know that the preservation method can have an impact on the cell types recovered from a given tissue, which preservation method is best for a cell type of interest needs to be determined empirically for each tissue type. Scientists need to be aware of the limitations of each sample type, the biases that sample preparation can introduce, and design their experiments according to their experimental goals.
Case Study: Reliable sample preparation uncovers potential therapeutic targets
An experiment to measure gene expression in tumor infiltrating leukocytes (TILs) between different tumors from patients
Using these experimental design considerations, we designed an experiment using scRNA-seq to identify potential therapeutic targets that facilitate T-cell infiltration into lung tumors. Fresh samples were obtained from patients to facilitate the study. Single-cell suspensions from fresh lung tumor tissue samples with matched normal resections from three different patients were prepared using the automated Singulator® Platform. Tumor-infiltrating lymphocytes (TILs) were immunomagnetically selected from the suspensions using the cell surface CD45 epitope and sequenced alongside normal cell and tumor cell libraries. Finally, the gene expression data of TILs isolated from tumors was compared to gene expression data from healthy T-cells in normal adjacent tissue to discover potential therapeutic targets.
The Singulator Platform generates reproducible and consistent single-cell suspensions
First, we confirmed the Singulator Platform’s performance, including the reliability of the Singulator Platform, to generate single-cell suspensions from biological replicates. This allowed us to estimate cellular composition differences in healthy vs cancerous tissues and simultaneously measure the gene expression profiles. In normal adjacent lung samples, the cell populations recovered demonstrate all of the expected cell types with the expected variation between different biological replicates (Figure 3, 4).
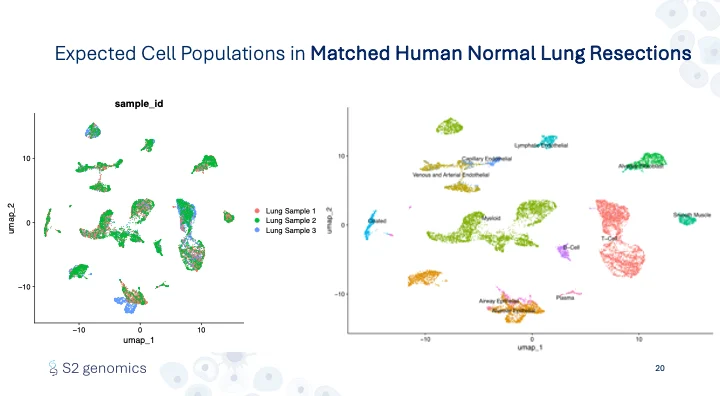
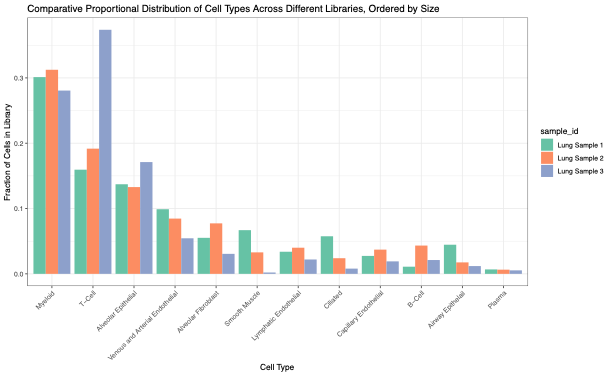
Across the three lung tumor samples – from three distinct types of non-small cell lung cancers containing differing cell type populations – gene expression profiles reflected an increase in cellular heterogeneity in tumors, and a unique cell profile for each tumor type (Figure 5, 6).
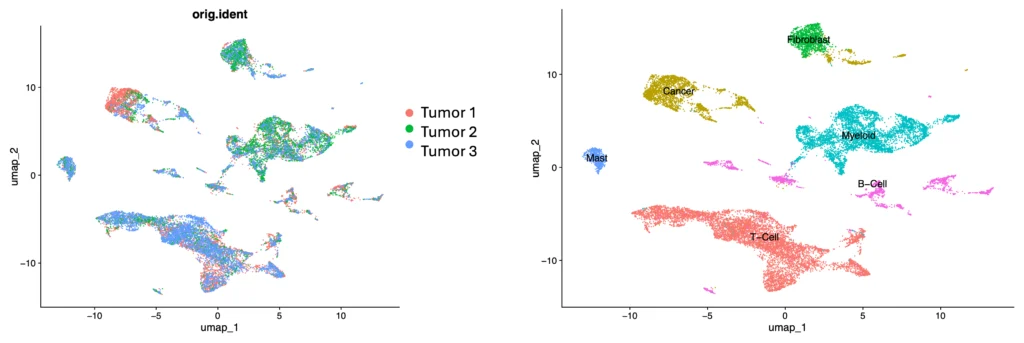
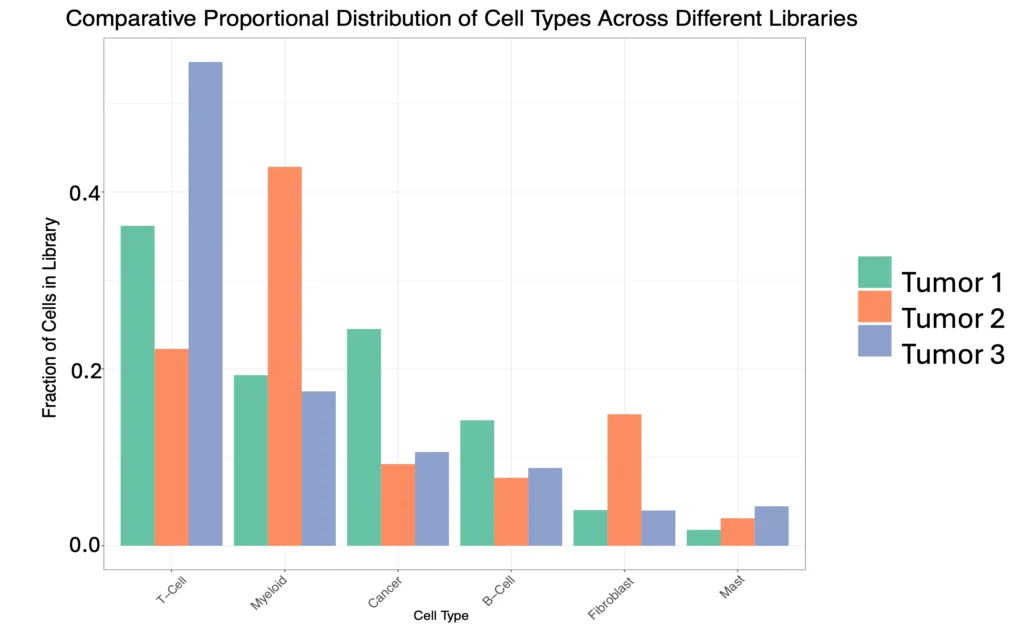
The Singulator Platform preserves cell surface epitopes
Next, the successful isolation of the TIL population from tumor samples was verified by comparing PTPRC (a marker of CD45-positive T-cells) gene expression profiles from the TIL samples and tumor samples (Figure 7). Analysis verified successful isolation of TILs, an indication that single-cell preparation preserved the CD45 surface epitope, enabling immunomagnetic separation of a targeted cell population from Singulator-prepared single-cell suspensions.
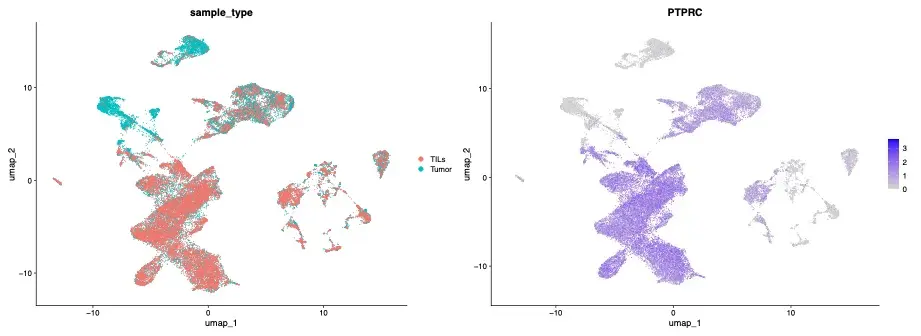
Identification of therapeutic targets
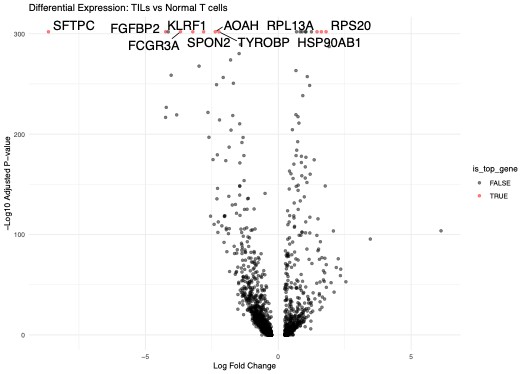
Finally, the gene expression profiles of T-cell populations from the TIL and normal human lung tissue samples (separated using CD3 gene expression) were compared to identify potential T-cell biomarkers that may facilitate tumor infiltration. Upon further investigation of the top ten differentially regulated genes, we discovered two of these genes, FCGR3A and HSP90AB1, had been previously identified in the literature and are under investigation to be therapeutic targets, validating the experimental design and reliability of the sample preparation (Figure 8).
The Automation Advantage in Reliable Sample Preparation
The Singulator Platform offers reproducible and precise sample preparation
The Singulator Platform offers unparalleled benefits for sample preparation in oncology research. Its automated system ensures reliability and minimizes variability, which is crucial for sensitive single-cell studies. By streamlining workflows, it reduces hands-on time and the potential for human error, while preserving the integrity of both cells and nuclei. The Platform’s flexibility supports a wide range of tissue types and preservation methods, making it an ideal solution for researchers aiming to achieve high-quality, consistent results in nuclei isolation, cell isolation, and single-cell dissociation. This reliability translates into more accurate data, saving time and money while facilitating breakthrough discoveries in tumor heterogeneity and targeted therapies.
Introducing the Singulator 200+
The upcoming Singulator 200+ automates the isolation of cells and nuclei from diverse tissue types, including FFPE samples, with minimal hands-on time. This Platform simplifies workflows, ensuring consistent, high-quality results for single-cell applications.
To learn more about the methods used to generate high-quality, single-cell and single-nuclei suspensions that capture tumor cell heterogeneity and enable targeted single-cell studies, watch our webinar, “Singulator Sessions II: Leveraging the Singulator Platform for Oncology Applications,” and read our App Note, “Single-Cell Sequencing of Tumor-Infiltrating Lymphocytes in Lung Cancer: A Comparative Analysis Using the Singulator™ Platform.”
Watch now or read the App Note here
References
- RNase Activity in Mouse Tissue: Classification, Hierarchy, and Methods for Control. Ambion TechNotes 12(3).
- Zhang, H., et al. Application of high-throughput single-nucleus DNA sequencing in pancreatic cancer. Nat Comm, 10 Feb 2023.
- Wu, H., et al. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol, 2019 Jan; 30(1):23-32.
- Kim, N. et al. Perspectives on single-nucleus RNA sequencing in different cell types and tissues. J of Path and Transl Med, 2023; 57:52-59.
- Abay, T. et al. Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing. Nature Genetics, 57,451-460 (2025).